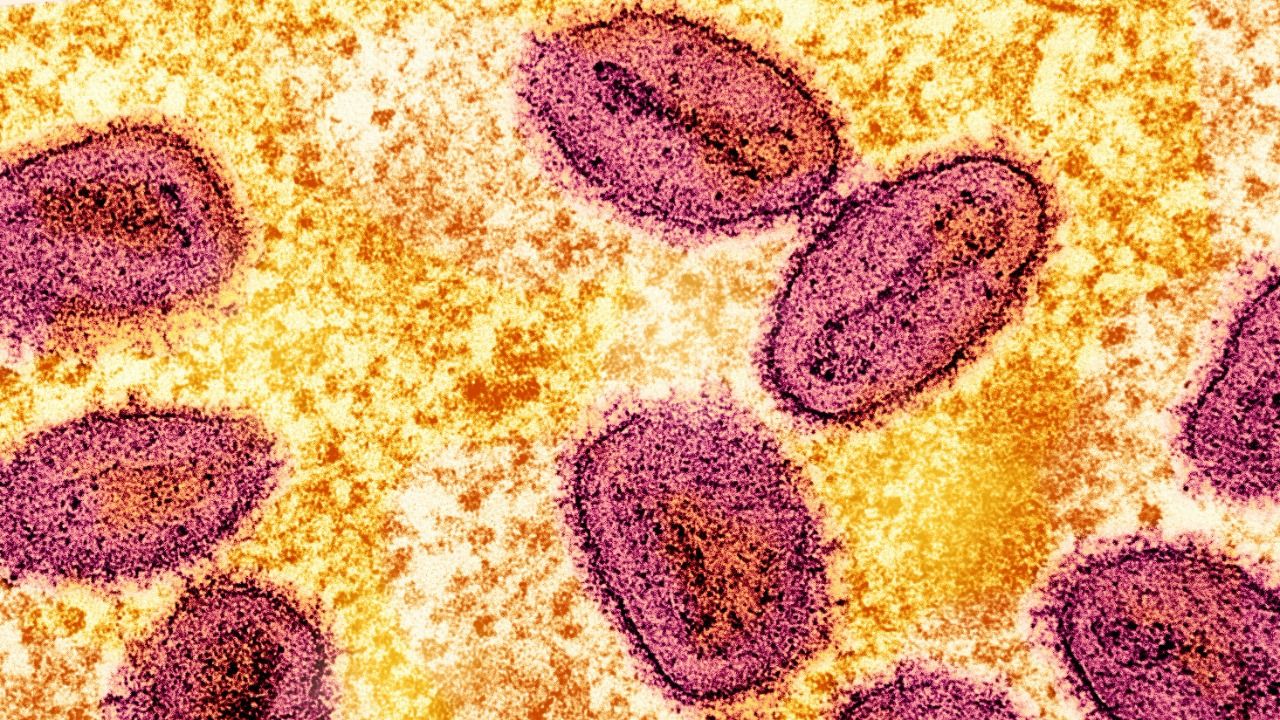
Artificial intelligence has just exposed a structural flaw in the mpox virus that scientists had not fully appreciated, and it sits on a protein that current vaccines barely touch. By zeroing in on this hidden vulnerability, researchers now see a path to vaccines and antibody drugs that are both more targeted and potentially more durable against future outbreaks. The discovery also shows how AI is starting to reshape the basic playbook for designing protection against poxviruses, from monkeypox to smallpox.
Why mpox still needed a better target
Mpox, often still called monkeypox, has been circulating in humans long enough to reveal the limits of the tools built for a different era. The main vaccines in use were originally designed with smallpox in mind, then repurposed when mpox surged, which means they focus on viral proteins that are important but not necessarily the most vulnerable. As the virus continues to affect communities across regions, the need for a more precise way to block infection has become harder to ignore.
Researchers working on mpox have been looking for a part of the virus that reliably provokes strong, protective antibodies in people who survive infection, yet is conserved enough that the virus cannot easily mutate around it. That is the context in which Dec findings about a little-known protein, identified with the help of artificial intelligence, have landed with such force. In multiple reports, Dec is cited as the moment when an AI-guided analysis of the mpox genome and protein structures pointed scientists toward a previously overlooked viral component that behaves like a pressure point for the immune system, a feature that existing vaccines do not fully exploit.
The AI-assisted hunt for a hidden protein
The breakthrough began with a simple but powerful idea: instead of starting from what scientists already believed were the key antigens, let AI search the entire viral proteome for patterns that human intuition might miss. Researchers fed structural and sequence data from the mpox virus into advanced models that can predict how proteins fold and how antibodies might latch onto them. The goal was to let algorithms surface candidates that looked unusually promising as antibody targets, even if they had been largely ignored in past vaccine work.
According to Dec reports, Researchers used AI to pinpoint a little-known monkeypox protein that provokes strong protective antibodies, a target that had not been at the center of earlier vaccine designs. When the team validated this AI-selected protein in laboratory experiments, they found that antibodies directed against it could neutralize the virus with striking potency. One detailed account explains that Researchers used AI to pinpoint a little-known monkeypox protein that provokes strong protective antibodies, and that the protein identified through AI analysis became the centerpiece of a new strategy for both vaccines and antibody therapies, a shift that would have been unlikely without computational help.
From survivors’ blood to a new weak spot
To understand which viral pieces really matter in the human body, scientists did not rely on AI alone. They started with people who had already faced down the virus. In one project, investigators collected blood from individuals who survived infection with monkeypox virus, then isolated the antibodies that their immune systems naturally produced. By mapping which viral proteins those antibodies recognized most strongly, they could see where the human immune response was already finding leverage.
Those real-world antibody profiles were then layered onto AI predictions to narrow the search. A report describing this approach notes that “We started with people who survived infection with monkeypox virus, isolated antibodies that they naturally produced” and then used those antibodies to guide the hunt for better ways to fight mpox, including targets that might be shared across poxviruses that are still impacting humans. In parallel, another Dec account explains that Scientists leveraged AlphaFold 3 to pinpoint a previously overlooked monkeypox protein that antibodies latch onto, using the structural predictions to understand exactly how those survivor-derived antibodies were binding and why that interaction was so effective.
What makes OPG153 different from classic vaccine targets
The protein that emerged from this combined AI and immunology effort, identified in the scientific literature as OPG153, stands apart from the viral components that older vaccines emphasize. Traditional smallpox-era vaccines tend to focus on surface proteins that are abundant and easy for the immune system to see, but those proteins are not always the ones that generate the broadest or most durable neutralizing response. OPG153, by contrast, appears to be a structural element that is both accessible to antibodies and functionally important to the virus, a combination that makes it a particularly attractive weak point.
One detailed summary of the work describes how the antigen-agnostic strategy led to broadly neutralizing antibodies targeting OPG153, a finding captured in the reference “Antigen-agnostic identification of poxvirus broadly neutralizing antibodies targeting OPG153” by Ida Paciel and colleagues. That reference, highlighted in a Dec report, underscores that the team did not begin with preconceived notions about which antigen to chase, but instead let data from human antibodies and AI-driven structural models converge on OPG153 as a standout. Another account notes that the study, which carries the DOI 10.1126/scitranslmed.aeb3840, was supported in part by the Welch Foundation, a reminder that this kind of deep structural work requires sustained investment as well as computational ingenuity.
How AI reshaped the vaccine design playbook
Once OPG153 was flagged as a prime target, the question became how to turn that insight into practical tools. Here again, AI played a central role. Instead of designing vaccine candidates by trial and error, researchers used models to predict which fragments of OPG153 would be most likely to trigger strong neutralizing antibodies, and how to present those fragments in a way that mimics the natural viral structure. That allowed them to rapidly iterate on potential vaccine constructs in silico before moving only the most promising designs into animal and cell-based testing.
One Dec report describes how an international team, working with artificial intelligence, made the first major step toward a new mpox vaccine by identifying and structurally characterizing this weak spot, a milestone captured in a piece that credits the effort to an international collaboration and notes that the lead author of the study relied on AI to guide the structural analysis. Another account frames the same advance as a major inroad toward a new and more effective way to fight monkeypox virus, explaining that Now, a major inroad towards a new and more effective way to fight monkeypox virus has been published, with the OPG153 target opening the door to vaccines and antibody therapies that are tuned specifically to this vulnerable site rather than spread thinly across less critical antigens.
From vaccine concept to antibody therapies
The implications of this discovery extend beyond preventive shots. Once scientists know that antibodies against OPG153 can broadly neutralize mpox, they can begin to design monoclonal antibody therapies that deliver those protective molecules directly. Such treatments could be used to protect people who are exposed during an outbreak, to treat those who are already infected, or to safeguard individuals who cannot mount a strong response to vaccination, such as some immunocompromised patients.
Reporting on the work emphasizes that the same AI-guided mapping that identified OPG153 is now informing the development of antibody cocktails that bind to different parts of the protein, reducing the chance that the virus can escape. One detailed summary notes that Now, a major inroad towards a new and more effective way to fight monkeypox virus has been published, and that the OPG153 target is being used to design antibody therapies to treat mpox or for use in high-risk settings. By focusing on a protein that appears conserved across multiple poxviruses, these antibodies may also offer a measure of cross-protection, a strategic hedge against future relatives of mpox that could spill over into humans.
AI as a tip-off for new drug targets
While OPG153 has captured much of the attention, the same AI-driven approach is also pointing scientists toward other parts of the mpox virus that could be vulnerable to drugs. Instead of looking only at surface proteins, researchers are scanning the entire viral machinery for enzymes and structural components that small molecules could disrupt. The idea is to let AI sift through thousands of possible viral functions and flag those that are both essential to the virus and structurally suited to binding by a drug.
One report describes how AI Tips Off Scientists to a New Drug Target to Fight and Treat Monkeypox Virus, detailing how computational models highlighted a specific viral protein that plays a central role in replication. That work, shared through Austin News, explains that the team used AI to predict the three-dimensional structure of the viral component they targeted, then identified pockets on its surface where a drug could lodge and disable its function. By treating AI as a tip-off system rather than a black box, the researchers were able to move quickly from prediction to experimental validation, turning an abstract model into a concrete therapeutic lead.
What this means for future outbreaks
The mpox work offers a glimpse of how outbreak response might look in the coming decade if AI-guided discovery becomes standard. Instead of waiting years to understand which viral proteins matter most, scientists could use structural prediction tools like AlphaFold 3 within weeks of sequencing a new pathogen, then cross-reference those predictions with antibody responses from early survivors. That combination, already demonstrated in mpox, could rapidly surface hidden weak spots that traditional approaches might overlook.
Several Dec accounts stress that the mpox findings are not just about one virus, but about a template for dealing with poxviruses more broadly, including smallpox. One detailed summary notes that Dec research on this AI-identified protein is expected to guide future efforts against smallpox, since the same or similar proteins appear in related viruses. Another report, which highlights Credit for the structural imagery to iStock and describes how With the help of artificial intelligence an international team made this first major step, underscores that the tools used here are general purpose. If the world faces another orthopoxvirus threat, the combination of AI structure prediction, survivor antibody mapping, and antigen-agnostic screening that led to OPG153 can be redeployed almost immediately.
The limits and next steps for AI-guided vaccines
For all the excitement, the mpox breakthrough is still at an early stage, and AI is not a magic wand. Models can predict promising targets, but they cannot replace the painstaking work of testing vaccine candidates in animals and eventually in people. There is also the risk that viruses will evolve under the pressure of new vaccines and therapies, forcing scientists to update their designs. The hope is that by focusing on structurally constrained proteins like OPG153, where mutations carry a high cost for the virus, AI can help pick targets that are harder for the pathogen to escape.
Researchers involved in the mpox work have been explicit that their antigen-agnostic approach depends on high quality experimental data as much as on algorithms. The reference to Antigen-agnostic identification of poxvirus broadly neutralizing antibodies targeting OPG153 by Ida Paciel and colleagues makes clear that the team validated AI predictions with detailed structural biology and immunology, not just computational scores. Another Dec account, which notes that the study’s DOI is 10.1126/scitranslmed.aeb3840 and that it was supported in part by the Welch Foundation, highlights the scale of the effort required to turn an AI hint into a fully characterized vaccine target. The next steps will involve translating that target into clinical candidates, tracking how the virus responds, and refining the models with each new round of data.
A new model for human–AI collaboration in virology
What stands out in the mpox story is not just the specific protein that AI helped uncover, but the way human expertise and machine prediction were woven together. Immunologists brought deep knowledge of how survivors’ antibodies behave, structural biologists interpreted complex protein folds, and AI specialists tuned models like AlphaFold 3 to the quirks of poxvirus proteins. Each group relied on the others to avoid blind spots, and the result was a target that none of them might have found as quickly on their own.
One Dec report on UT researchers describes how they used AI to develop better ways to fight mpox, starting from survivor antibodies and moving through a pipeline that integrated computational and experimental steps. Another account, which explains that AI Tips Off Scientists to a New Drug Target to Fight and Treat Monkeypox Virus, shows the same pattern on the therapeutic side, with AI acting as a scout rather than a replacement for laboratory work. Taken together with the structural studies that identified OPG153 and the antigen-agnostic strategy documented by Ida Paciel, these efforts sketch a new model for virology in which AI is embedded from the first days of an outbreak through the final stages of vaccine and drug design. If mpox is any guide, that model could make the difference between chasing a virus and getting ahead of it.
More from MorningOverview